Despite the fundamental role networks play in how scientists understand the dynamics and properties of complex systems, reconstructing networks from large-scale experimental data is a challenge.
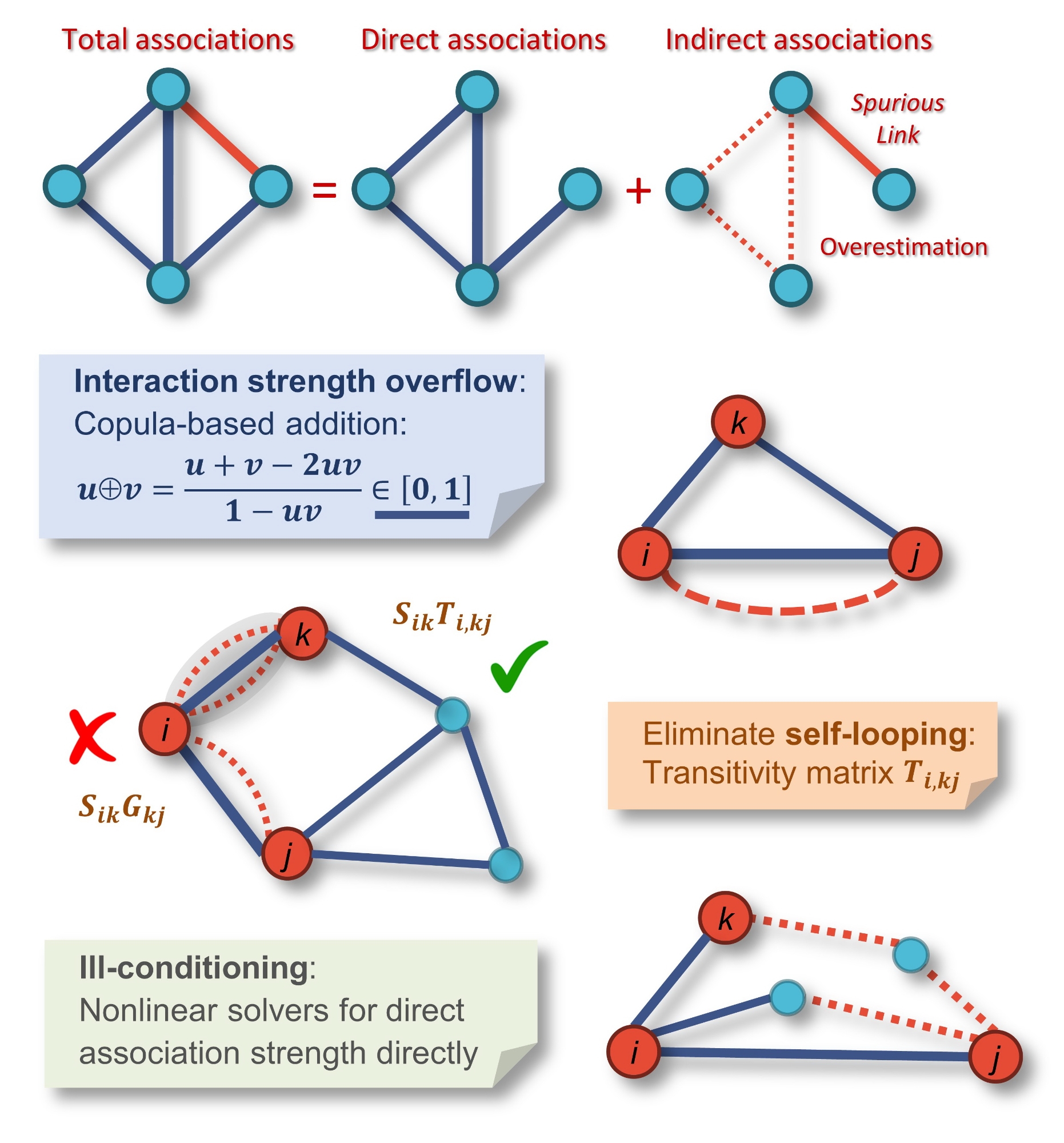
In systems biology and microbial ecology – the study of microbes in the environment and their interactions with each other – the challenges of reconstructing these networks can be compounded by difficulty unraveling direct and indirect interactions, or the ability of one element in a system to impact another, either with or without direct interaction.
Jizhong Zhou, director of the Institute for Environmental Genomics at the University of Oklahoma, is leading a research team at IEG, in collaboration with Mary Firestone from the University of California at Berkeley, to demonstrate the effectiveness of a new conceptual framework for disentangling direct and indirect relationships in association networks. Zhou is also a George Lynn Cross Research Professor in the Dodge Family College of Arts and Sciences and an adjunct professor in the Gallogly College of Engineering at OU.
Naijia Xiao, a research scientist at IEG, is the first author of the paper describing this work, published in the journal Proceedings of the National Academy of Sciences.
“By tackling several mathematical challenges, this study provides a conceptual framework for disentangling direct and indirect relationships in association networks,” Zhou said. “The application of our iDIRECT (Inference of Direct and Indirect Relationships with Effective Copula-based Transitivity) framework to synthetic gene expression and microbial community data demonstrates that the framework is a powerful, robust and reliable tool for network inference. It will greatly enhance our capability to discern network interactions in various complex systems and will allow scientists to address research questions which could not be approached previously.”