Experiment Summary and Publications
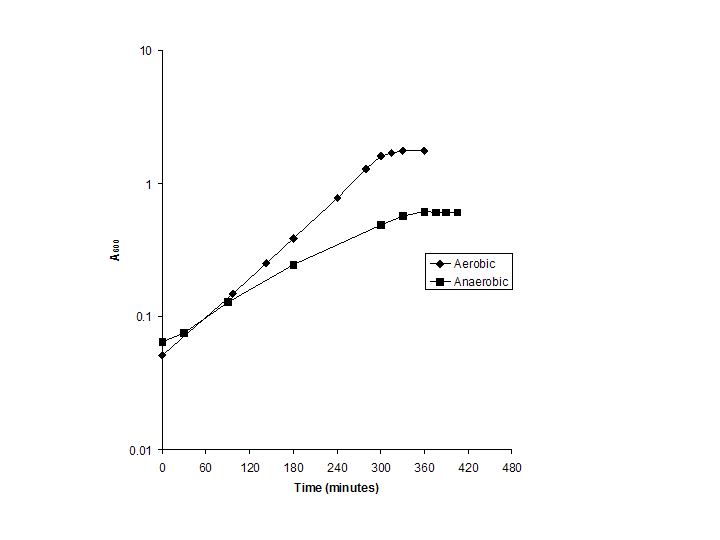
| Time (min) |
Aerobic |
Timepoint |
Time (min) |
Anaerobic |
| 0 |
0.051 |
1 |
0 |
0.065 |
| 97 |
0.148 |
2 |
30 |
0.0755 |
| 143 |
0.252 |
3 |
90 |
0.129 |
| 180 |
0.388 |
4 |
180 |
0.247 |
| 240 |
0.775 |
5 |
300 |
0.4905 |
| 280 |
1.285 |
6 |
330 |
0.568 |
| 300 |
1.61 |
7 |
360 |
0.613 |
| 315 |
1.7 |
8 |
375 |
0.6105 |
| 330 |
1.765 |
9 |
390 |
0.61 |
| 360 |
1.765 |
10 |
405 |
0.605 |
Experimental Design
Culture conditions. E. coli MG1655 (CGSC #6300) was cultured in 1 liter of morpholinepropanesulfonic acid (MOPS) minimal medium containing 2.0 g/liter of glucose (Neidhardt et al., 1974) in a 2 liter Biostat B fermentor. The temperature was maintained at 37 degrees C and pH was kept constant at 7.2 by the addition of 2 M NaOH. For the aerobic culture, the dissolved oxygen level was maintained above 20% of its saturation level by adjusting the agitation speeds in the range of 400-600 rpm with fixed 1 liter/min air flow rate. For the anaerobic culture, the medium contained 35 mM Na H2CO3 and was sparged with a mixture of nitrogen and carbon dioxide gases (90:10) at a fixed 1 liter/min flow rate. Growth was monitored by measuring the optical density (OD) at 600nm. The culture continued growing until entering stationary phase upon exhaustion of carbon and nitrogen. RNA samples were isolated at appropriate intervals during the experiment by pipeting into ice-cold RNAlater™ (Ambion, Austin, TX) followed by purification using an RNeasy™ Mini Kit (Qiagen, Valencia, CA). The RNA samples were labeled by first strand cDNA synthesis. Labeled targets were hybridized to DNA arrays (Panorama E. coli Gene Arrays, Sigma Genosys Biotechnologies, Inc., The Woodlands, TX). The hybridized arrays were scanned by phosphorimaging at a pixel density of 100 microns (10,000 dots/cm2) with a STORM 820 PhosphoImager (Molecular Dynamics, Sunnyvale, CA) following exposure to a Kodak Storage Phosphor Screen GP (Eastman Kodak Co., Rochester, NY) for 24 hrs. The array membranes were consecutively hybridized, stripped, and rehybridized.Spot-finding and quantitation. Image analysis software (ArrayVision, Imaging Research, Inc.) was used for spot-finding and quantitation of the E. coli Panorama arrays. The raw spot intensities were represented in a row-column format and exported into Microsoft Excel spreadsheets for further analysis, or as comma-delimited files (.csv) for upload to the database. Raw data from each experimental replicate were analyzed in Excel workbooks containing manually executed macros written in Visual Basic, or the data were processed in the database. The first step in the analysis associates the array coordinate for each spot with a unique spot number, the gene name, and related gene annotation. On the membrane arrays there are two spots for each gene, and these were treated as separate determinations. The raw data were normalized by expressing spot intensities as a percentage of the sum of all of the gene-specific spot intensities. The second step in the analysis applies the student t-test to determine the probability that the average of the experimental replicates is significantly different from the average of the control replicates . The P values (derived from the student t-test) for the normalized and natural log transformed data were calculated. The third step calculates relative gene expression between conditions by introducing a threshold value, chosen to be representative of the limit of detection of expressed genes (usually the 500th lowest expressed gene), and then calculating the ratio of the experimental/control expression levels such that genes that are more highly expressed in the experimental condition are given a positive value, and genes that are more highly expressed in the control condition are given a negative value.
Control for ratio calculation. Timepoint 2 provided the early log phase control values for the aerobic growth curve data set, as originally published (Chang, et al., 2002). The data are also analyzed with control values obtained by averaging of all replicates for timepoints 1-4. This more robust strategy for establishing the control values was also used for analysis of the aerobic and anaerobic time series data..
Data Legend
Aerobic Growth Data Set (Timepoint 2 control;
Excel: 3.1 Mb)
TGC_2_conpct: average normalized spot intensity
for control timepoint 2; values expressed as a percentage of the
sum of all of the gene-specific spot intensities
TGC_10_tstpct: average normalized spot intensity
for test timepoint 10
TGC_10_V_2_logratio: log10 ratio of timepoint 10
vs. timepoint 2
TGC_10_V_2_pln: P value for the corresponding log
ratio calculated from the normalized, natural log transformed data
Aerobic Growth Data Set (Timepoint
1-4 averaged control; Excel: 3.4 Mb)
TGC_1-4_conpct : average normalized spot intensity
for control timepoints 1-4; values expressed as a percentage of
the sum of all of the gene-specific spot intensities
TGC_1_tstpct: average normalized spot intensity
for test timepoint 1
TGC_1_logratio: log10 ratio of timepoint 1 vs.
control average timepoints 1-4
TGC_1_pln: P value for the corresponding log ratio
calculated from the normalized, natural log transformed data
AGTC_2_conpct: average normalized spot intensity for control timepoint 2; values expressed as a percentage of the sum of all of the gene-specific spot intensities
AGTC_10_tstpct: average normalized spot intensity for test timepoint 10
AGTC_10_V_2_logratio: log10 ratio of timepoint 10 vs. timepoint 2
AGTC_10_V_2_pln: P value for the corresponding log ratio calculated from the normalized, natural log transformed data
Anaerobic Growth Data Set (Timepoint
1-4 averaged control; Excel: 3.4 Mb)
AGTC_1-4_conpct : average normalized spot intensity
for control timepoints 1-4; values expressed as a percentage of
the sum of all of the gene-specific spot intensities
AGTC_1_tstpct: average normalized spot intensity
for test timepoint 1
AGTC_1_logratio: log10 ratio of timepoint 1 vs.
control average timepoints 1-4
AGTC_1_pln: P value for the corresponding log ratio
calculated from the normalized, natural log transformed data
Aerobic vs. Anaerobic Comparison
(Timepoints 1-6 averaged Excel: 1.5Mb)
Aer_pct: average normalized spot intensity for
aerobic timepoints 1-6; values expressed as a percentage of the
sum of all of the gene-specific spot intensities
Anaer_pct: average normalized spot intensity for
anaerobic timepoints 1-6
Anaer/Aer_LOGRATIO: log10 ratio of Anaerobic average
timepoints 1-6 vs. Aerobic average timepoints 1-6
Anaer/Aer_PLN: P value for the corresponding log
ratio calculated from the normalized, natural log transformed data
Aer_STDDEV: Standard deviation of Aerobic timepoints
1-6
Anaer_STDDEV: Standard deviation of Anaerobic timepoints
1-6
E. coli Gene Expression Database (Oracle) Interface
Raw Data File Available on request