Using R Packages on OSCER Systems
Table of Contents
What version of R is available on OSCER resources?
Click here to find what version of R is installed and available on OSCER systems.
You can always get the latest information by following the steps below:
- Log into the OSCER system
- Type module avail R
- Scroll down the list until you see the listing for R (similar to the highlighted image below)
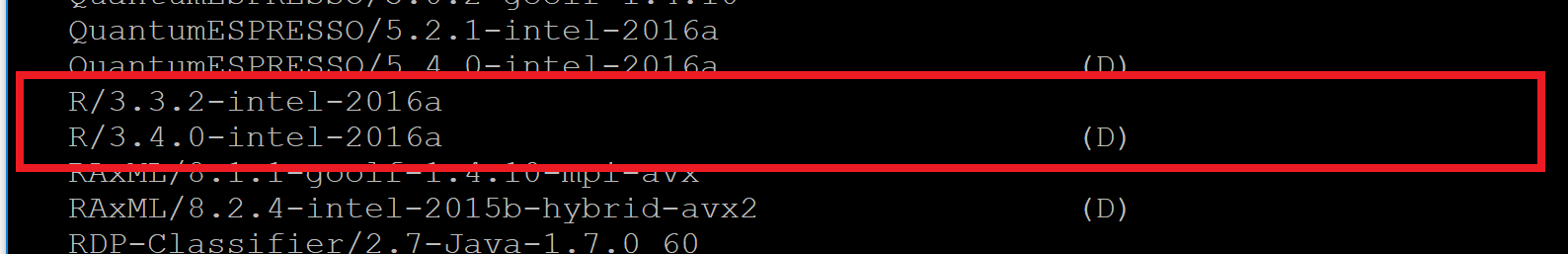
You can pick whichever version of R works best for your project, but should try to use the latest version available for best results. That is currently R/4.0.3-foss-2020b, but that will change.
What R packages are available on OSCER resources?
OSCER maintains several R packages that can be used without the need to install a local package. Follow the steps below to get the most up-to-date list:
- Log into the OSCER system
- Load the module for the version of R you need by typing (for example): module load R/4.0.3-foss-2020b
- Then start an R session by typing: R
- To obtain the list of R packages, type: library()
Starting R
- Log into the OSCER system
- Load the module for the version of R you need by typing (for example): module load R/4.0.3-foss-2020b
- Then start an R session by typing: R
This loads R in interactive mode and will place you inside the R console. This is the place where you would install additional R packages, and run small R scripts (less than 3 minutes in duration).
NOTE: Any computational intensive work should be submitted as a job. Please refer to Running R in a Job.
Installing Additional R Packages
This section guides you through the process of installing R packages locally without root access on OSCER systems.
We highly recommend looking through the pre-installed packages to see if the package you want is already installed on OSCER systems. To do this, please refer to What R packages are available on OSCER resources?
The follow steps will guide you through the process of installing local R packages in your home directory. Please note that we do not recommend installing R packages in your scratch folder because of our autodelete policy.
In this example, we will install the package ACSWR from https://cran.r-project.org/web/packages/available_packages_by_name.html
- Log into the OSCER system
- Load the module for the version of R you need by typing(for example): module load R/4.0.3-foss-2020b
- Start an R session by typing: R
- To install the ACSWR package, type: install.packages(c("ACSWR"))
- You will be prompted with the following message:
Installing package into ‘/usr/lib64/R/library’
(as ‘lib’ is unspecified)
Warning in install.packages(c("ACSWR")) :
'lib = "/usr/lib64/R/library"' is not writable
Would you like to use a personal library instead? (y/n) - Type y and press Enter. This tells R that you will be installing the package in a local folder that you have access to (your home directory).
- Next, you will be prompted with the following message:
Would you like to create a personal library
~/R/x86_64-redhat-linux-gnu-library/3.3
to install packages into? (y/n) - Type y and press Enter. This will create the folder /R/x86_64-redhat-linux-gnu-library/3.3 in your home directory.
- Next, you'll get the following message asking you to choose an install source:
--- Please select a CRAN mirror for use in this session ---
HTTPS CRAN mirror
1: 0-Cloud [https] 2: Algeria [https]
3: Australia (Canberra) [https] 4: Australia (Melbourne 1) [https]
5: Australia (Melbourne 2) [https] 6: Australia (Perth) [https]
7: Austria [https] 8: Belgium (Ghent) [https]
9: Brazil (PR) [https] 10: Brazil (RJ) [https]
11: Brazil (SP 1) [https] 12: Brazil (SP 2) [https]
13: Bulgaria [https] 14: Canada (BC) [https]
15: Canada (MB) [https] 16: Canada (NS) [https]
17: Chile 1 [https] 18: Chile 2 [https]
19: China (Beijing) [https] 20: China (Hefei) [https]
21: China (Guangzhou) [https] 22: China (Lanzhou) [https]
23: China (Shanghai 1) [https] 24: China (Shanghai 2) [https]
25: Colombia (Cali) [https] 26: Czech Republic [https]
27: Denmark [https] 28: East Asia [https]
29: Ecuador (Cuenca) [https] 30: Ecuador (Quito) [https]
31: Estonia [https] 32: France (Lyon 1) [https]
33: France (Lyon 2) [https] 34: France (Marseille) [https]
35: France (Montpellier) [https] 36: France (Paris 2) [https]
37: Germany (Erlangen) [https] 38: Germany (Göttingen) [https]
39: Germany (Münster) [https] 40: Greece [https]
41: Iceland [https] 42: India [https]
43: Indonesia (Jakarta) [https] 44: Iran [https]
45: Ireland [https] 46: Italy (Padua) [https]
47: Japan (Tokyo) [https] 48: Japan (Yonezawa) [https]
49: Korea (Seoul 1) [https] 50: Korea (Ulsan) [https]
51: Malaysia [https] 52: Mexico (Mexico City) [https]
53: New Zealand [https] 54: Norway [https]
55: Philippines [https] 56: Serbia [https]
57: Singapore (Singapore) [https] 58: Spain (A Coruña) [https]
59: Spain (Madrid) [https] 60: Sweden [https]
61: Switzerland [https] 62: Taiwan (Chungli) [https]
63: Turkey (Denizli) [https] 64: Turkey (Mersin) [https]
65: UK (Bristol) [https] 66: UK (London 1) [https]
67: USA (CA 1) [https] 68: USA (IA) [https]
69: USA (IN) [https] 70: USA (KS) [https]
71: USA (MI 1) [https] 72: USA (NY) [https]
73: USA (OH) [https] 74: USA (OR) [https]
75: USA (TN) [https] 76: USA (TX 1) [https]
77: Vietnam [https] 78: (HTTP mirrors) - Type 1 and press Enter.
- At this point, the system will attempt to download the source package and install it in your home directory.
- Upon completion, you have successfully installed the ACSWR package in the following location in your home directory: /R/x86_64-redhat-linux-gnu-library/3.3/ACSWR
Running R in a Job
The sample code below shows how to submit an R job via the batch script. If you are not familiar with OSCER's job scheduler or submitting batch scripts, please refer to the following tutorials first:
- http://www.ou.edu/oscer/support/running_jobs_schooner
- http://www.ou.edu/oscer/support/running_jobs_schooner#nonparallel
Steps to submit an R job:
Step 1: Create a sample R job script called helloworld.r and insert the following line of code:
print("hello world")
Step 2: Create a batch submission file called r_batch.sh and insert the following lines of code:
#!/bin/bash
#
#SBATCH --partition=normal
#SBATCH --ntasks=1
#SBATCH --mem=1024
#SBATCH --output=r_output_%J.txt
#SBATCH --error=r_error_%J.txt
#SBATCH --time=12:00:00
#SBATCH --job-name=jobname
#SBATCH --mail-user=youremailaddress@yourinstitution.edu
#SBATCH --mail-type=ALL
#SBATCH --chdir=/home/yourusername/directory_to_run_in
#
#################################################
module load R/4.0.3-foss-2020b
Rscript helloworld.r > output.txt
In summary, the batch script asks for 1 CPU core along with 1024MB of memory for 12 hours. If your job expeccts to run on multiple cores in parallel, please specify that in '--ntasks=' instead. If it can run on all cores in a node (currently 20 or 24 on Schooner copmute nodes), please replace '--ntasks=1' with '--exclusive', to request the entire node, and prevent other jobs from running there at the same time.
Once the job starts, it runs the commands:
module load R/4.0.3-foss-2020b
Rscript helloworld.r > output.txt
on whatever compute node the job is running on. The first command prepares to run R. And the second actually runs the R script helloworld.R and places the output in the file output.txt. You can change these to whatever values are appropriate for you.
Note: Any large output should go to your /scratch space as your home directory is modest (currently 20GB). /scratch is never backed up and is subject to file purging/cleaning of older files (files older than 2 weeks).
Step 3: Submit the batch job by typing sbatch r_batch.sh
Step 4: Upon successful completion of the job, you will find three new files in your directory: output.txt, r_output_<SLURM_jobID>.txt and r_error_<SLURM_jobID>.txt. The '%J' will be replaced by the SLURM jobID, therefore avoiding overwriting files from older jobs.
Step 5: If the job ran successfully you should see the results in output.txt. If your R job produces other output files, they will be created in that directory as well.
Resources
Additional support references can be found here:
- http://www.rexamples.com/
- https://www.datamentor.io/r-programming/examples
- https://people.eng.unimelb.edu.au/aturpin/R/
- http://www.win-vector.com/blog/2016/01/parallel-computing-in-r/
- http://dept.stat.lsa.umich.edu/~jerrick/courses/stat701/notes/parallel.html
- https://cran.r-project.org/web/views/HighPerformanceComputing.html
- https://www.rdocumentation.org/packages/parallel/versions/3.5.0

